Research
We employ genetic, biochemical, and genomic approaches to manipulate and study proteins that deposit, remove, and decode chromatin modifications to reveal how they regulate gene expression and maintain genome stability.
Chromatin modifications alter the landscape of the genome and play critical roles in facilitating DNA replication, transcription, recombination, and repair through mechanisms that remain poorly understood. Currently, the genome-wide patterns of many chromatin modifications are known and, in many cases, such modifications can be correlated with a measurable outcome, like gene expression. However, it remains unclear how specific patterns of chromatin modifications are established, how they are recognized by the relevant cellular machinery, and how they are translated into the desired outcome. Focusing largely on DNA and histone methylation, my laboratory seeks to understand how chromatin modifications facilitate gene regulation and contribute to genome stability via the silencing of transposons and the coordination of DNA repair. To this end, we are addressing the following questions using the plant model Arabidopsis thaliana: (1) How are specific patterns of DNA methylation established and modulated? (2) How does DNA methylation affect gene expression? (3) How do chromatin modifications facilitate DNA repair? As both the DNA methylation and DNA repair pathways are highly conserved, our work takes advantage of the speed and genetic malleability of a model system while maintaining a high level of relevance to both agriculture and human health.
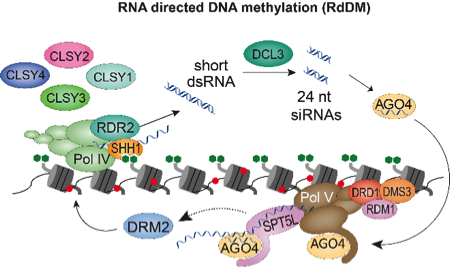 (1) How are specific patterns of DNA methylation established and modulated?
(1) How are specific patterns of DNA methylation established and modulated?
We recently identified the CLSY family as the major factors controlling the locus-specific deposition of DNA methylation via the RNA-directed DNA methylation pathway. We now aim to leverage further characterization of these factors to determine the mechanisms underpinning more complex aspects of DNA methylation, including the maintenance of methylation homeostasis, the generation of tissue and cell-type specific methylation patterns, and the modulation of the epigenome by environment interactions.
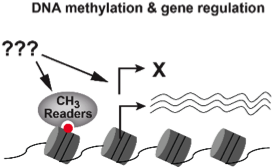 (2) How does DNA methylation affect gene expression?
(2) How does DNA methylation affect gene expression?
Currently very little is understood about how DNA methylation regulates gene expression. Recently, we identified several protein complexes that contain functional methyl-DNA binding proteins. Characterization of theses complexes revealed roles in promoting, rather than suppressing, gene expression. As these factors act largely downstream of DNA methylation, understanding how they function will provide mechanistic insights into how DNA methylation influences gene expression.
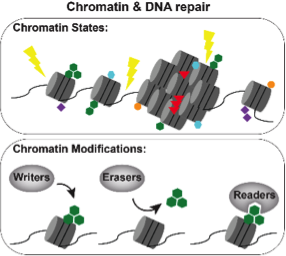 (3) How do chromatin modifications facilitate DNA repair?
(3) How do chromatin modifications facilitate DNA repair?
During DNA repair, chromatin constitutes both a physical barrier to overcome, as the repair machinery must gain access to the damaged DNA, and a molecular platform for the recruitment of DNA repair proteins. This process, like transcriptional regulation, is influenced by a variety of chromatin modifications. Recently, we generate a transcriptional network of the DNA damage response and we are now applying our expertise in chromatin biology to understand how specific chromatin modifications influence the process of DNA repair to promote genome stability.